

The Aging Research Centre (ARC) is developing an Automated Imaging Microscope System (AIMS) to enable researchers to view the three-dimensional cellular organization of a region of tissue from a series of microscope slides. AIMS employs a computer-controlled microscope that can move a microscope slide along the X and Y axes, so that the field of view can be moved in equal increments. The microscope also has an autofocus mechanism. A CCD camera captures images, one field of view at a time. Using a magnification of 400 power, a typical slide contains 25,000 fields of view; perhaps 7000 of those fields contain something of interest. By using the thousands of individually captured images, the system is able to reconstruct a very high-resolution image of the entire slide.
AIMS analyzes each of the thousands of images and identifies and stores the following attributes for each cell it was instructed to identify: X,Y and Z coordinates for each desired cell type, amount of each stain in the cell, type of each stain in the cell, color of each stain in the cell and type of cell.
When AIMS has processed multiple layers of a tissue block, the system will be able to reconstruct a three-dimensional map of the cells in that block. In an experiment where there are two groups of animals and the researcher wishes to compare tissues from the two groups, AIMS will be able to compare three-dimensional structures as well as cell counts.
The system has two very different functions: acquisition and analysis. When we designed the system, we started on both ends simultaneously, attempting to analyze images captured from other sources, then working on our own data. Because we hope for the system to be used by biology researchers, not UNIX gurus, a decent user interface is one of the primary goals. We chose Tcl/Tk for the ease with which a GUI can be developed. The easily modifiable prototypes are useful. Various commands, written in C (but with Tcl/Tk interfaces) are used to control physical devices.
Physically, the system consists of an optical microscope, a CCD camera and three stepper motors hooked to the parallel port. The stepper motors move a microscope slide around the stage under computer control; basically, we have a 60,000bpi scanner.
The stepper motors are driven by Darlington-pair transistors (see http://www.doc.ic.ac.uk/~ih/doc/stepper/ for a discussion on stepper-motor control). This allows for great control, at the expense of hefty timing requirements. In the Linux kernel, we can find floppies driven every three to eight milliseconds. Empirically, given the load on our motors, we can sometimes move more than twice a second. We've hooked them up to the parallel port. The parallel port has eight data lines and four control lines which can be used for output. This is just sufficient to control three stepper motors at four controls/motor. If we needed more motors, we would have to use a different type of controller. Figure 1 shows the microscope looking at the focus mechanism. Figure 2 shows the circuit driving the motors. It uses two ULN2003A chips and is powered by a spare computer power supply.

Figure 1. The Microscope
We need to do a lot of moving, so speed is very important. Using nanosleep seems the simplest, if not the best, alternative. Combined with real-time priority, this causes the motors to move with a nice smooth hum. The other alternative, using the real-time clock at 2048KHz, doesn't allow as precise a control over speed. The major problem with this approach is the way nanosleep handles its delays. A busy-wait prevents any other task from running. RTLinux does seem like a better solution, although we haven't investigated that yet. The older usleep call is a poor choice, as it has 10ms granularity.

Figure 2. Circuit that Drives Motors
The greatest problem is figuring out the speed to move the motor. The speed at which a stepper motor is capable of moving depends on the load placed on it, and this load varies depending on the friction in the stage. A speed that works at one location may not work at all at another. Empirical experimentation seems necessary.
We can move in three dimensions. Not only can we view the entire slide, we can change the focus. A jury-rigged solution seems to work—move up and down, and pick the image with the most detail. We assume the one with the most detail is the most focused. Technically, “detail” is based on a “busyness” function. For each pixel, find the difference in intensity between each of its neighbors, then sum the absolute value of those differences.
Because this project started before Linux 2.2, which has frame grabbers built into the kernel, we are using the Matrox meteor frame grabber, which has support at www.gnofn.org/~marksu/meteorman.html. The mvid program which comes with the driver was a useful starting point. We integrated it with Tcl/Tk. This allows us to make snapshots, view real-time video at variable frame rates and sizes over our network and get measures such as “how dark is it?” or “how much detail is there?” by directly accessing frame-buffer memory.
Real-time video is useful for manual focus to check whether autofocus is working, for setting boundaries of the scan, and of course, for just joking around by taking pictures of staff members. The “meteor” driver sends out a signal whenever a new image is available. If we're ready, we will send the image out using XPutImage and XSync. If the previous image isn't done, we ignore the frame entirely.
While shape is important, size and color are simpler to use as heuristics. We take a single image, then use sliders to select the colors which we consider to be a cell. If it is big enough and the right color, it must be a cell. This isn't a very sophisticated technique; it isn't much of a refinement over “thresholding”, where anything sufficiently dark is counted.
Currently, we use Tcl/Tk to select the ranges of RGB color which will be allowed. In the future, it may be useful to select regions in HSV color space.
The simplicity of the algorithm means cells can be counted “on the fly”; during the scan, the algorithm is performed on each field of view. The cells on the boundaries are counted multiple times, but we know where the boundaries are and can ignore them.
It would theoretically be possible to do this job without any computer at all. A technician could look at each slide, 0.2 mm at a time, and count every cell he saw. Looking at 2mm by 2mm sections, this would require exhaustive work for the 100-odd fields covered by a typical mouse hypothalamus. Fatigue could introduce bias. It would be easy to count a given marginal case one way when wide awake and another when tired—but people are good at image processing, computers aren't. People make mistakes when they are tired; computers make mistakes all the time. Still, even if absolute numbers are biased, we hope that relative numbers will still show useful differences.
There are currently two interfaces to the physical “scanner”: one for grabbing an overview (see Figure 3) of the entire slide, at 25 bits per inch (i.e., the microscope's objective is moved 1mm at a time, and the average color at that point is saved), and another for grabbing a specified region on the slide. In the second case, because of the low speed of directory listings (ls takes quite a bit of time if there are 2000 files), a directory is created for every column scanned. Figure 4 shows the interface used to scan in a rectangular region. The user can use the cursor keys to move the slide, and then select the boundaries.
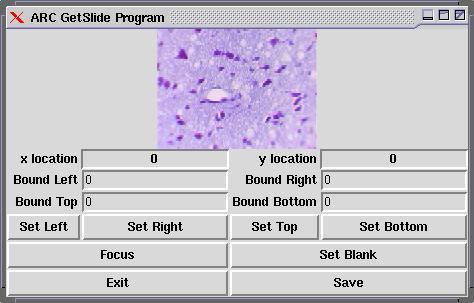
Figure 4. Scan Interface
One planned refinement is to scan in only those areas which we think may have useful content. If locations (x,y), (x+1mm,y), (x,y+1mm), (x+1mm,y+1mm) are all blank, it is reasonable (given the size of our samples) to ignore (x+0.5mm, y+0.5mm).
The optimal refinement would be to store only the regions which actually have useful content. In our case, we are interested in only the hypothalamus. An empty area is near this, which could conceivably be automatically recognized; if so, we could discard thousands of frames of less-important data.
It would be nice to store the entire slide in a standard image format such as JPEG or TIFF, but for some reason, 12,5000x50,000-pixel images are difficult to process at 24 bits per pixel (18GB per image seems a little excessive). Storing each frame individually using JPEG uses 10-50KB per frame; more for detailed ones, less for blanks. If only images with useful detail are saved, it should get under 650MB/slide, in which case each slide might be stored on a CD-ROM.
Given the non-standard format, we will need a way to view individual segments on a slide. A multi-resolution display tool uses preprocessed images from the slide to create a mosaic, which shows multiple frames at the same time. By saving each frame at multiple resolutions, we do not need to decompress hundreds of JPEG images, only to throw 9999/10,000ths of the detail away.
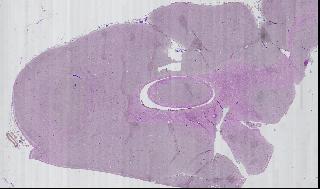
Figure 5
Using this tool, we can zoom out to view the entire slide, zoom in on a specific region of a single sample, then view a specific field of vision. Figure 5 is a mosaic of one portion of one slide.
Refinements would allow integration with the counting algorithm. By drawing a border around a region of interest, counts and densities of cells in that region could be displayed.
When successive sections of the same tissue sample are placed in a known sequence, those successive sections can be merged to form a single three-dimensional image of the original tissue. Distortions in the sample are likely to cause some difficulty, but standard feature-recognition techniques should be able to compensate.
A further refinement is possible. With cells ~25 microns in size, 4 micron slices span several cells. By staining successive sections with different factors, it is possible to determine several different facts about each cell.
The Automated Imaging Microscope System (AIMS) will be used by my colleague Lee R. McCook at the University of California at Berkeley. He will use the system to compare the 3-D cellular densities in the hypothalamus of normal mice and calorically restricted mice. I have downloaded his Ph.D. proposal to the Aging Research Centre's web site at www.arclab.org/linux/leephd.html.
ARC has two research facilities in North America: the first is located in Berkeley, California and the second in Waterloo, Ontario, Canada. The near-term objectives of the Aging Research Centre are to conduct experiments that shed light on the underlying mechanisms that cause the aging process to occur and to develop tools which will help researchers better understand why humans and other organisms undergo this process. Our long-term objective is to use the information that has been accumulated on this subject and develop intervention procedures that will dramatically slow down the aging process. The aging of an organism is a very complex and multifaceted process that encompasses many disciplines in biology such as neuroendocrinology, histology, genetics, enzymology, biochemistry, molecular biology and so on. At this time, there is approximately 100GB of information in journals and books that relates to the aging process, and more and more information is being added every day to this body of knowledge. One of the tools we are developing will allow a researcher to see this massive quantity of data via a computer using graphical and interactive methods in order to represent this information in a more comprehensible and coherent manner.
The other major tool is the focus of this article: an Automated Imaging Microscope System to be used for an experiment at the University of California at Berkeley. It will be used to study the effects of calorie deprivation on neural tissue. Mice on calorie-reduced diets often live longer, and this may be based on changes in the hypothalamus.